Today, as the biomedical community is exploring all options to address COVID-19, the ability to review and synthesize all available data is critical for identifying potential therapies. But this immense focus on COVID-19 is yielding a rapidly growing body of new information for scientists to read and manually curate — all in addition to the existing body of data on SARS and other coronaviruses. Reading all of the research would take thousands of hours, and finding connections between numerous entities can be daunting.
The solution is a map that surfaces all connections from source biomedical documents. This requires:
- expertly curated biomedical data at large scale, and
- modern AI technologies, like the ubiquitous personal assistants that help us in our everyday lives.
Recent advances in AI technologies enable computers to scan billions of documents and identify critical connections based on the meaning of the biomedical text. Data scientists refer to this meaning as semantics in order to distinguish it from legacy text analytics approaches that merely map the syntax of word patterns. These semantic connections can then be visualized to quickly identify insights and link those insights back to every supporting document. When working with biomedical text data, AI can replace thousands of hours of searching within data silos ranging from PubMed to clinical trials to preprint servers. This ability to move beyond lengthy manual research is precisely what scientists need to accelerate a COVID-19 treatment.
This modern approach to parsing scientific data using AI is at the core of tellic graph, a biomedical knowledge graph that is currently used at several biopharma and academic centers to expedite research. As part of the response to COVID-19, tellic assembled the data related to COVID-19 disease in a cloud-based tool called graph.C19.
At the center of graph.C19 are COVID-19, SARS and other coronavirus diseases — to enable researchers to parse the growing body of biomedical data on COVID-19 as well as learn from prior experience with coronaviruses. From those key diseases, all known connections to drugs, phenotypes, genes and variants are included in the dataset, as well as additional connections for some of those entities. Users can start with any or all three diseases and branch out to explore all available knowledge.
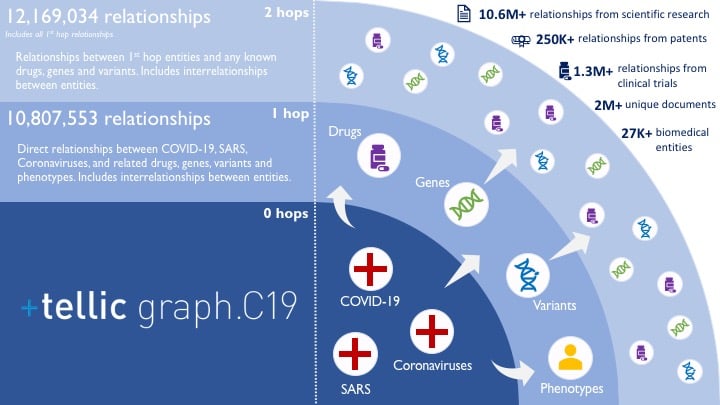
With this interactive tool, researchers can instantaneously find all relationships between biomedical entities in a query. With manual search and review approaches, however, this work would take substantial effort and time commitment, plus rare but pivotal connections could be missed entirely.
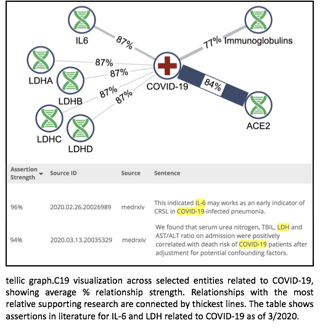 In sum, included within tellic graph.C19 are 12.2 million relationships among 27,525 biomedical entities supported by data from 2 million source documents. As expected, top studied nodes from COVID-19 are the virus receptor ACE2, immunoglobulins and IL6. However, it is interesting to note that the strongest assertions in literature related to COVID-19 are IL6, and the lactate dehydrogenase genes (LDHA-D), highlighting their role as well-characterized biomarkers of advanced disease.
In sum, included within tellic graph.C19 are 12.2 million relationships among 27,525 biomedical entities supported by data from 2 million source documents. As expected, top studied nodes from COVID-19 are the virus receptor ACE2, immunoglobulins and IL6. However, it is interesting to note that the strongest assertions in literature related to COVID-19 are IL6, and the lactate dehydrogenase genes (LDHA-D), highlighting their role as well-characterized biomarkers of advanced disease.
We invite scientists to register for free access to tellic graph.C19, and explore the connections relevant to COVID-19 research.

